About NetPyNE
NetPyNE (Networks using Python and NEURON) is a Python package to facilitate the development, simulation, parallelization, analysis, and optimization of biological neuronal networks using the NEURON simulator.
Although NEURON already enables multiscale simulations ranging from the molecular to the network level, using NEURON for network simulations requires substantial programming, and often requires parallel simulations. NetPyNE greatly facilitates the development and parallel simulation of biological neuronal networks in NEURON for students and experimentalists. NetPyNE is also intended for experienced modelers, providing powerful features to incorporate complex anatomical and physiological data into models.
For a more detailed overview of NetPyNE see:
Our NetPyNE article in the journal eLife
Our NetPyNE tutorial from CNS*2022
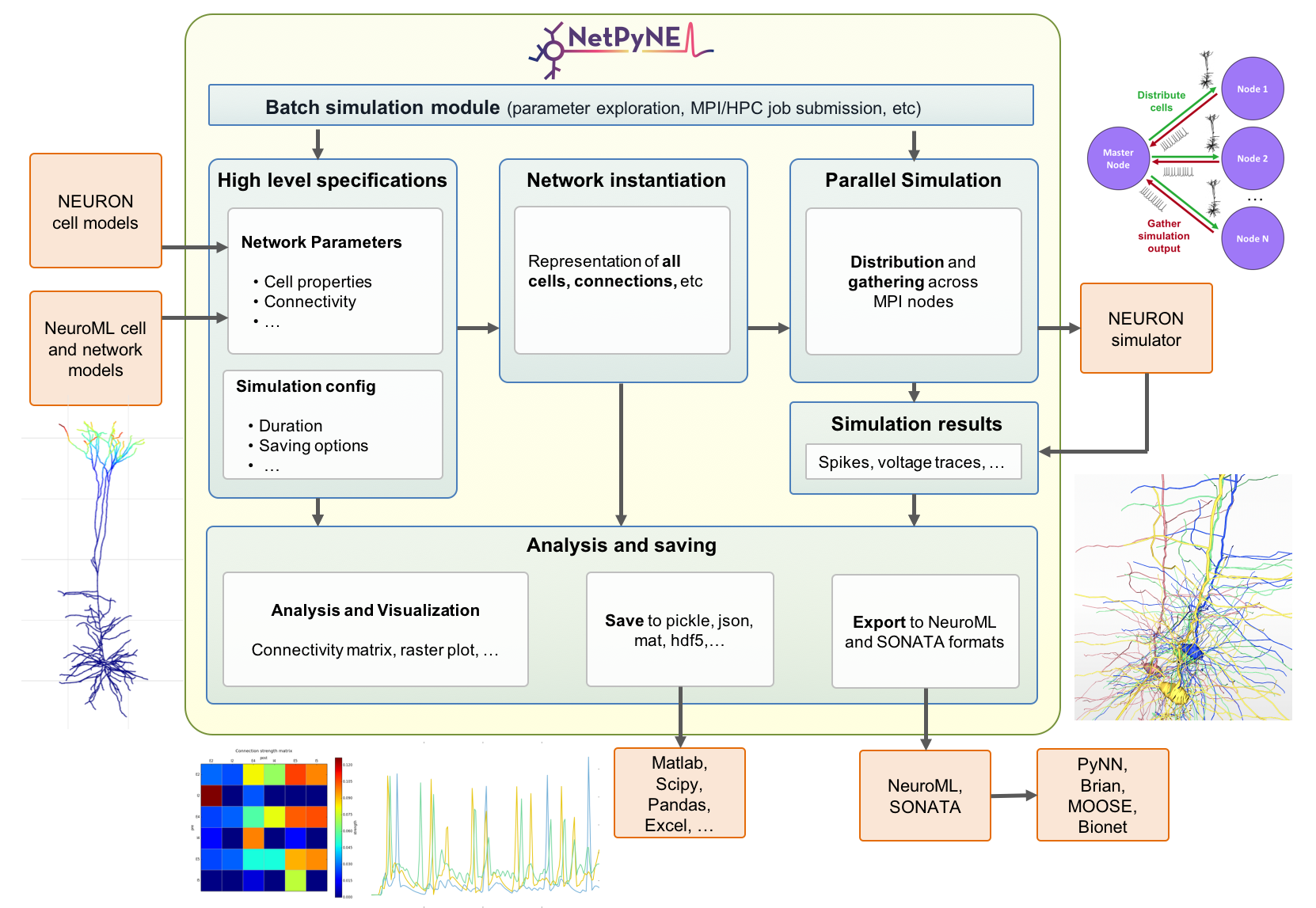
Major Features
Converts a set of high-level specifications into a NEURON network model
Specifications are provided in a simple, standardized, declarative Python-based format
Can easily define:
Populations: cell type and model, number of neurons or density, spatial extent, …
Cell properties: morphology, biophysics, implementation, …
Synaptic mechanisms: time constants, reversal potential, implementation, …
Stimulation: spike generators, current clamps, spatiotemporal properties, …
Connectivity rules: conditions of pre- and post-synaptic cells, different functions, …
Simulation configuration: duration, saving and analysis, graphical output, …
Reaction-diffusion (RxD): species, regions, reactions, …
Cell properties highlights:
Import existing HOC and Python defined cell models into NetPyNE format
Readily change model implementation e.g., from Hodgkin-Huxley multicompartment to Izhikevich point neuron
Combine multiple cell models into hybrid networks for efficient large-scale networks
Connectivity rules highlights:
Flexible connectivity rules based on pre- and post-synaptic cell properties (e.g., cell type or location)
Connectivity functions available: all-to-all, probabilistic, convergent, divergent, and explicit list
Can specify parameters (e.g., weight, probability or delay) as a function of pre/post-synaptic spatial properties, e.g., delays or probability that depend on distance between cells or cortical depth
Can specify subcellular distribution of synapses along the dendrites, and will be automatically adapted to the morphology of each model neuron.
Can easily add learning mechanisms to synapses, including STDP and reinforcement learning
Generates NEURON network instance ready for MPI parallel simulation
Takes care of cell distribution
Handles gathering of data
Analysis and plotting of network and simulation output:
Raster plot of all cells or populations
Spike histogram of all cells, populations, or single cells
Intrinsic cell variable plots (voltages, currents, conductances)
Local field potential (LFP) calculation and plots (time-resolved and power spectra)
Connectivity matrix at cell or population level (weights, number of connections, efficiency, probability, …)
2D representation of network cell locations and connections
3D shape plot with option to include color-coded variables (e.g., number of synapses)
Normalized transfer entropy and spectral Granger Causality
Facilitates data sharing:
Can save/load high-level specs, network instance, simulation configuration, and simulation results.
Multiple formats supported: pickle, Matlab, JSON, CSV, HDF5
Can export/import to/from NeuroML and SONATA, standardized formats for neural models
Batch simulations:
Easy specification of parameters and range of values to explore in batch simulations
Pre-defined, configurable setups to automatically submit jobs in multicore machines (bulletin board) or supercomputers (SLURM or PBS Torque)
Analysis and visualization of multidimensional batch simulation results
Current usage:
Used to develop models of many different brain regions and phenomena. See [full list of models](www.netpyne.org/models)
Integrated with the Human Neocortical Neurosolver to add flexibility to its cortical model
Used by Open Source Brain to run parallel simulation of NeuroML-based NEURON models
Available to run simulations on XSEDE supercomputers via the Neuroscience Gateway
Questions, suggestions, and contributions
Two kinds of contributions are solicited: 1. direct contributions to NetPyNE code described below. 2. Ancillary code contributions.
NetPyNE is open-source and available at github.com/Neurosim-lab/netpyne.
For questions or suggestions please use the NetPyNE forum , the NEURON+NetPyNE forum or submit an issue to GitHub.
Contributions are gladly welcomed! Please fork the repository and make a Pull Request with your changes. See our Contributors Guide for more details.
For further information please contact: salvadordura@gmail.com.
Code of conduct
This project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.
We as members, contributors, and leaders pledge to make participation in our community a harassment-free experience for everyone, regardless of age, body size, visible or invisible disability, ethnicity, sex characteristics, gender identity and expression, level of experience, education, socio-economic status, nationality, personal appearance, race, religion, or sexual identity and orientation.
We pledge to act and interact in ways that contribute to an open, welcoming, diverse, inclusive, and healthy community.
Please read the full Code of Conduct.
Publications
About NetPyNE
Dura-Bernal S, Suter B, Gleeson P, Cantarelli M, Quintana A, Rodriguez F, Kedziora DJ, Chadderdon GL, Kerr CC, Neymotin SA, McDougal R, Hines M, Shepherd GMG, Lytton WW. NetPyNE: a tool for data-driven multiscale modeling of brain circuits. eLife 2019;8:e44494 , 2019.
Lytton WW, Seidenstein AH, Dura-Bernal S, McDougal RA, Schurmann F, Hines ML. Simulation neurotechnologies for advancing brain research: Parallelizing large networks in NEURON. Neural Computation, 2016.
Dura-Bernal S, Suter BA, Quintana A, Cantarelli M, Gleeson P, Rodriguez F, Neymotin SA, Hines M, Shepherd GMG, Lytton WW. NetPyNE: a GUI-based tool to build, simulate and analyze large-scale, data-driven network models in parallel NEURON. Society for Neuroscience (SfN), 2018.
Dura-Bernal S, Suter BA, Neymotin SA, Shepherd GMG, Lytton WW. Modeling the subcellular distribution of synaptic connections in cortical microcircuits. Society for Neuroscience (SFN), 2016.
Dura-Bernal S, Suter BA, Neymotin SA, Kerr CC, Quintana A, Gleeson P, Shepherd GMG, Lytton WW. NetPyNE: a Python package for NEURON to facilitate development and parallel simulation of biological neuronal networks. Computational Neuroscience (CNS), 2016.
Gleeson P, Marin B, Sadeh S, Quintana A, Cantarelli M, Dura-Bernal S, Lytton WW, Davison A, Silver RA. A set of curated cortical models at multiple scales on Open Source Brain. Computational Neuroscience (CNS), 2016.
Dura-Bernal S, Suter BA, Neymotin SA, Quintana AJ, Gleeson P, Shepherd GMG, Lytton WW. Normalized cortical depth (NCD) as a primary coordinate system for cell connectivity in cortex: experiment and model. Society for Neuroscience (SFN), 2015.
Make use of NetPyNE
Terra, H.d.C.B., Borges, F.d.S., Moraes, M.F.D., Cota, V.R. Implementation of Intra and Extracellular Nonperiodic Scale-Free Stimulation in silico for the NEURON Simulator. Communications in Computer and Information Science, vol 1519. Springer, Cham. 2021. doi:https://doi.org/10.1007/978-3-031-08443-0_8
Ying Y, Yubo F, Songan H, Qingyun W. Optogenetic stimulation of primary motor cortex regulates beta oscillations in the basal ganglia: A Computational study Communications in Nonlinear Science and Numerical Simulation, vol 117. 2023. doi:https://doi.org/10.1016/j.cnsns.2022.106918
Hasegan D, Deible M, Earl C, D’Onofrio D, Hazan H, Anwar H, Neymotin SA. Multi-timescale biological learning algorithms train spiking neuronal network motor control bioRxiv 2021.11.20.469405, 2021. doi: https://doi.org/10.1101/2021.11.20.469405.
Bryson A, Berkovic SF, Petrou S, Grayden DB. State transitions through inhibitory interneurons in a cortical network model. PLoS Comput Biol 17(10): e1009521, 2021. doi: https://doi.org/10.1371/journal.pcbi.1009521.
Pimentel1 JM, Moioli RC, de Araujo MFP, Ranieri CM, Romero RAF, Broz F and Vargas PA. Neuro4PD: An Initial Neurorobotics Model of Parkinson’s Disease. Front. Neurorobot. 15:640449, 2021. doi: 10.3389/fnbot.2021.640449.
Volk, V.L., Hamilton, L.D., Hume, D.R. et al. Integration of neural architecture within a finite element framework for improved neuromusculoskeletal modeling. Scientific Reports 11, 22983, 2021. doi: https://doi.org/10.1038/s41598-021-02298-9.
Ranieri CM, Moioli RC, Vargas P, Romero RAF, A Neurorobotics Approach to Behaviour Selection based on Human Activity Recognition, arXiv:2107.12540 [cs.RO], 2021. https://arxiv.org/abs/2107.12536.
Ranieri CM, Pimentel JM, Romano MR, Elias LA, Romero RAF, Lones MA, Araujo MFP, Vargas PA, Moioli RC, A Data-Driven Biophysical Computational Model of Parkinson’s Disease Based on Marmoset Monkeys, IEEE Access, vol. 9, pp. 122548-122567, 2021. doi: https://doi.org/10.1109/ACCESS.2021.3108682.
Metzner C, Mäki-Marttunen T, Karni G, McMahon-Cole H, Steuber V. The Effect of Alterations of Schizophrenia-Associated Genes on Gamma Band Oscillations, bioRxiv 2020.09.28.316737, 2020. doi: https://doi.org/10.1101/2020.09.28.316737.
Gao P, Graham J, Zhou W, Jang J, Angulo S, Dura-Bernal S, Hines ML, Lytton WW, and Antic S Local Glutamate-Mediated Dendritic Plateau Potentials Change the State of the Cortical Pyramidal Neuron, Journal of Neurophysiology (In Press). 2020.
Sivagnanam S, Gorman W, Doherty D, Neymotin S, Hovhannisyan H, Fang S, Lytton W, Dura-Bernal S Simulating large-scale models of brain neuronal circuits using Google Cloud Platform, Practice and Experience in Advanced Research Computing, PEARC2020, 2020.
Neymotin, S.A., Daniels, D.S., Caldwell, B., McDougal, R.A., Carnevale, N.T., Jas, M., Moore, C.I., Hines, M.L., Hamalainen, M. and Jones, S.R., Human Neocortical Neurosolver (HNN), a new software tool for interpreting the cellular and network origin of human MEG/EEG data, eLife, 9, p.e51214, 2020. doi: https://elifesciences.org/articles/51214 .
Lytton WW, Sherif MA, Dura-Bernal S, Neymotin S. Multiscale modeling of a “mental” disease, Virtual Physiological Human Conference, VHP2020, 2020.
Dura-Bernal S, Neymotin SA, Suter BA, Shepherd GMG, Lytton WW. Multiscale dynamics and information flow in a data-driven model of the primary motor cortex microcircuit, bioRxiv 201707, 2019. doi: https://www.biorxiv.org/content/10.1101/201707v3.
Gerkin, R.C., Birgiolas, J., Jarvis, R.J., Omar, C. and Crook, S.M.. NeuronUnit: A package for data-driven validation of neuron models using SciUnit, bioRxiv, p.665331, 2019.
Gleeson P, Cantarelli M, Quintana A, Earnsah M, Piasini E, Birgiolas J, Cannon RC, Cayco- Gajic A, Crook S, Davison AP, Dura-Bernal S, et al. Open Source Brain: a collaborative resource for visualizing, analyzing, simulating and developing standardized models of neurons and circuits, Neuron, 10.1016/j.neuron.2019.05.019, 2019. doi: https://www.cell.com/neuron/fulltext/S0896-6273(19)30444-1.
Dai K, Hernando J, Billeh JN, Gratiy SL, Planas J, Davison AP, Dura-Bernal S, Gleeson P, Devresse A, Gevaert M, King JG, Van Geit WAH, Povolotsky AV, Muller E, Courcol J-D, Arkhipov A . The SONATA Data Format for Efficient Description of Large-Scale Network Models, bioRxiv, 625491 [Preprint] doi: https://www.biorxiv.org/content/10.1101/625491v2. Under review in PLoS Computational Biology. 2019
Dura-Bernal S, Neymotin SA, Suter BA, Shepherd GMG, Lytton WW Long-range inputs and H-current regulate different modes of operation in a multiscale model of mouse M1 microcircuits, bioRxiv 201707, 2018. doi: https://www.biorxiv.org/content/10.1101/201707v3.
Dissertations:
Volk, V. Integrated Neuromusculoskeletal Modeling Within a Finite Element Framework to Investigate Mechanisms and Treatment of Neurodegenerative Conditions, Boise State University, 2022.
Conference abstracts:
Dura-Bernal S, Griffith EY, Barczak A, O’Connell MN, McGinnis T, Anwar H, Lytton WW, Lakatos p, Neymotin SA. Biophysically-detailed multiscale model of macaque auditory thalamocortical circuits reproduces physiological oscillations. Computational Neurosience (CNS), 2020.
Griffith EY, Dura-Bernal S, Barczak A, O’connell MN, Mcginnis TM, Lytton WW, Lakatos P, Neymotin SA. Data-driven model of auditory thalamocortical system rhythms. Society for Neuroscience (SfN), 2019.
Doherty DW, Dura-Bernal S, Lytton WW. Computer models of mouse area M1 show avalanches for full model and subcircuits defined by layer or cell type. Society for Neuroscience (SfN), 2019.
Dura-Bernal S, Menzies RS, McLauchlan C, van Albada SJ, Kedziora DJ, Neymotin SA, Lytton WW, Kerr CC. Effect of network size on computational capacity. Computational Neuroscience (CNS), 2016.
Romaro C, Araujo Najman F, Dura-Bernal S, Roque AC. Implementation of the Potjans-Diesmann cortical microcircuit model in NetPyNE/NEURON with rescaling option. Computational Neuroscience (CNS), 2018.
Rodriguez F. Dentate gyrus network model. Computational Neuroscience (CNS), 2018.
Cite NetPyNE
Spreizer S, Senk J, Rotter S, Diesmann M, Weyers, NEST Desktop, an Educational Application for Neuroscience eNeuro 11 November 2021, 8 (6) ENEURO.0274-21.2021, 2021. doi: http://doi.org/10.1523/ENEURO.0274-21.2021.
Amsalem, O., Eyal, G., Rogozinski, N., Gevaert, M., Kumbhar, P., Schürmann, F. and Segev, I., An efficient analytical reduction of detailed nonlinear neuron models. Nature Communications, 11(1), pp.1-13, 2020. doi: https://www.nature.com/articles/s41467-019-13932-6.
Billeh, Y.N., Cai, B., Gratiy, S.L., Dai, K., Iyer, R., Gouwens, N.W., Abbasi-Asl, R., Jia, X., Siegle, J.H., Olsen, S.R. and Koch, C.,. Systematic integration of structural and functional data into multi-scale models of mouse primary visual cortex. Neuron (In Press) NEURON-D-19-01027, 2020. doi: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3416643.
Tran, H., Ranta, R., Le Cam, S. and Louis-Dorr, V., Fast simulation of extracellular action potential signatures based on a morphological filtering approximation. Journal of Computational Neuroscience, pp.1-20, 2020. doi: https://doi.org/10.1007/s10827-019-00735-3.
Gast, R., Rose, D., Salomon, C., Möller, H.E., Weiskopf, N. and Knösche, T.R.. PyRates—A Python framework for rate-based neural simulations. PLoS ONE, 14(12), 2019. doi: https://doi.org/10.1371/journal.pone.0225900.
Tejada J, Roque AC, Conductance-based models and the fragmentation problem: A case study based on hippocampal CA1 pyramidal cell models and epilepsy Epilepsy & Behavior, 106841, 2019. doi: https://doi.org/10.1016/j.yebeh.2019.106841.
Kuhl E, Alber M, Tepole BA, Cannon WR, De S, Dura-Bernal S, Garikipati K, Karniadakis GE, Lytton WW, Perdikaris P, Petzold L. Multiscale modeling meets machine learning: What can we learn? arXiv:1911.11958. doi: https://doi.org/10.1007/s11831-020-09405-5. [Preprint]. Under review in Computer Methods in Applied Mechanics and Engineering. 2019
Alber M, Buganza A, Cannon W, De S, Dura-Bernal S, Garikipati K, Karmiadakis G, Lytton W, Perdikaris P, Petzold L, Kuhl E. Integrating Machine Learning and Multiscale Modeling: Perspectives, Challenges, and Opportunities in the Biological, Biomedical, and Behavioral Sciences. Nature Partner Journals (npj) Digital Medicine, 2, 115, 2019. doi: https://www.nature.com/articles/s41746-019-0193-y.
See here an updated list from Google Scholar.
Courses
Future
June 2022: Building and simulating brain circuit models on Google Cloud, Google Office, NYC (delayed)
Past
Dec 20201: Minicurso - Modelagem Computacional em NetPyNE usando Google Cloud Platform- Simpósio do PPGNCG (Brazilian course in portuguese)
July 2021: CNS’21 Building biophysically detailed neuronal models from molecules to networks with NEURON and NetPyNE.
May 2021: Online 2-week Course: Multiscale modeling of brain circuits using NetPyNE.
July 2020: CNS*2020 Building mechanistic multiscale models: from molecules to networks using NEURON and NetPyNE, online conference
January 2020: VIII Latin American School on Computational Neuroscience (LASCON), Institute of Mathematics and Statistics, University of Sao Paulo, Brazil
July 2019: CNS*2019 Tutorial organizer and lecturer, Building biophysically detailed neuronal models: from molecules to networks, Barcelona.
May 2019: Workshop on Multiscale Network Modeling, Brown University.
May 2019: Principles of Computational Neuroscience, Sassari University, Sardinia.
June 2018: NEURON Summer Course, Emory University, Atlanta.
July 2018: CNS/*2018 Multiscale Modeling from Molecular to Large Network Level, CNS/*2018, Seattle.
January 2018: VII Latin American School on Computational Neuroscience (LASCON), Institute of Mathematics and Statistics, University of Sao Paulo, Brazil
July 2017: Bernstein Computational Neuroscience Conference, Multiscale Modeling and Simulation, Gottingen.
Current funding
National Institutes of Health (NIH), National Insititute of Biomedical Imaging and Bioengineering (NIBIB) U24 EB028998: “Dissemination of a tool for data-driven multiscale modeling of brain circuits”, Period: 2019-2024; Amount: $1,171,482; PI: Salvador Dura-Bernal
Governance structure
Major decisions about NetPyNE are made by the steering committee, guided by the Project roadmap and the Code of conduct. The committee includes members from a diverse range of institutions, positions and backgrounds.
The current steering committee consists of the following members (in alphabetical order):
Salvador Dura-Bernal (Assistant Professor, State University of New York Downstate; Research Scientist, Nathan Kline Institute for Psychiatric Research)
Padraig Gleeson (Principal Research Fellow, University College London)
Joe W. Graham (Research Scientist, State University of New York Downstate)
Erica Y. Griffith (Graduate Student, State University of New York Downstate)
Michael Hines (Senior Research Scientist, Yale University)
Cliff C. Kerr (Senior Research Scientist, Institute for Disease Modeling)
William W. Lytton (Distinguished Professor, State University of New York Downstate; Kings County Hospital)
Robert A. McDougal (Assistant Professor, Yale University)
Samuel A. Neymotin (Research Scientist, Nathan Kline Institute for Psychiatric Research)
Benjamin A. Suter (Postdoctoral Fellow, Institute of Science and Technology Austria)
Subhashini Sivagnanam (Principal Computational and Data Science Research Specialist, San Diego Supercomputing Center)
Membership in the steering committee is a personal membership. Affiliations are listed for identification purposes only; steering committee members do not represent their employers or academic institutions.
Project roadmap
The five-year project roadmap (2019-2023) includes four large categories: quality control, development of new features, GUI extension, and dissemination and community engagement. The main targets for each category, and the estimated period
Quality control: robustness, reliability and reproducibility
2019-2021: Reliability - Test existing features, particularly recently added ones (RxD, subcellular connectivity, distributed saving, parameter optimization) such that they perform their intended function under all valid conditions and inputs.
2020-2022: Robustness and error handling - Ensure the tool is able to cope with erroneous inputs and errors during execution. Improved tool robustness will include input validation, exception handling and informational messages.
2022-2023: Reproducibility - Ensure simulation results are reproducible across the most common platforms, including different versions of operating systems, Python, NEURON, MPI library; and HPC platform setup (eg XSEDE/NSG, Google Cloud Platform).
Development of new features:
2020-2021: Macroscopic scale modeling - Extend the framework to support macroscale data (e.g. MRI, EEG, MEG) and models (e.g. mean field models), thus linking this scale to the underlying circuit, cellular and molecular mechanisms.
2021-2022: Machine learning analysis methods - Incorporate ML methods (e.g. clustering, dimensionality reduction, and deep learning) to explore and optimize large parameter spaces and analyze neural data.
2022-2023: Reverse engineering of networks - Infer high-level compact network connectivity rules (generative model) from the full connection information of biological network models, using statistical (e.g. Bayesian inference) and graph theoretical analysis.
GUI extension: Extension of the graphical user interface (GUI), essential to engage new users and make the tool accessible to experimentalists, clinicians and students.
2019-2020: Web-based multi-user deployment - Will allows users to build models and run simulations through a web browser over the internet, making the tool publicly available to the global research community.
2019-2022: Incorporating missing components - Currently only accessible programmatically: RxD, subcellular connectivity, complex stimulation and parameter optimization (only grid search).
2021-2022: Dynamic interactive plots - Improving plots by replacing the current static images with modern interactive and dynamic plots that facilitate understanding of complex and large datasets.
2022-2023: Visualization of large networks - Improving performance to enable 3D visualization and manipulation of large-scale networks of detailed neurons (currently limited to a few hundred neurons).
Dissemination and community engagement: We will implement complementary dissemination and engagement strategies to train and attract users and developers:
2019-2020: Online documentation - Updated and comprehensive online documentation covering all the tool components, options and modes of usage, with examples, so both beginner and advanced users can fully exploit the tool.
2019-2023: Workshops/tutorials - Organized at neuroscience conferences to engage potential users by providing an overview of the tool functionalities and benefits.
2020-2022: Online interactive tutorials - Will enable new users to receive training at their own pace through multimedia-rich step-by-step instructions that can be executed interactively (e.g. via GUI or Jupyter Notebook).
2020-2023: Annual 3-day in-person course - Will provide in-depth training to researchers/clinicians who could then teach tool usage at their labs or institutions.
2020-2023: Annual Hackathon - Will train and engage developers, promoting long-term, sustainable, collaborative development.